Machine-learning model can distinguish antibody targets
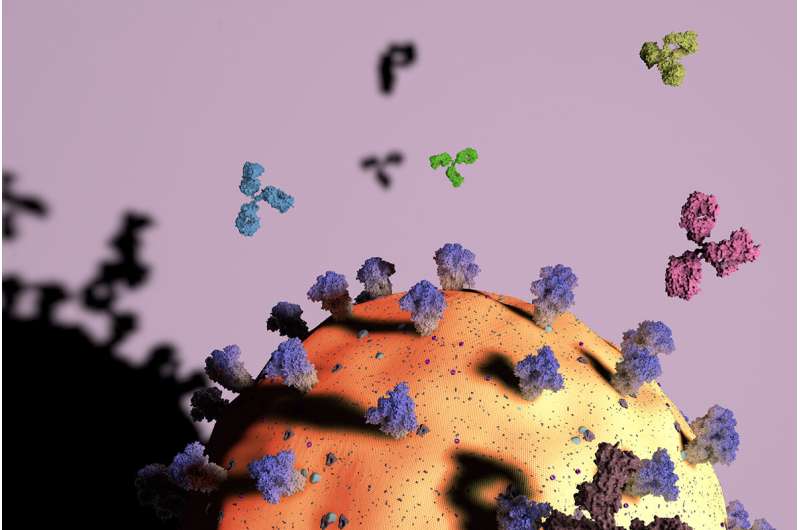
A new study shows that it is possible to use the genetic sequences of a person's antibodies to predict what pathogens those antibodies will target. Reported in the journalImmunity, the new approach successfully differentiates between antibodies against influenza and those attacking SARS-CoV-2, the virus that causes COVID-19.
"Our research is in a very early stage, but this proof-of-concept study shows that we can use machine learning to connect the sequence of an antibody to its function," said Nicholas Wu, a professor of biochemistry at the University of Illinois Urbana-Champaign who led the research with U. of I. biochemistry Ph.D. student Yiquan Wang; and Meng Yuan, a staff scientist at Scripps Research in La Jolla, California.
With enough data, scientists should be able to predict not only thevirusan antibody will attack, but which features on the pathogen the antibody binds to, Wu said. For example, an antibody may attach to different parts of thespike proteinon the SARS-CoV-2 virus. Knowing this will allow scientists to predict the strength of a person's immune defense, as some targets of a pathogen are more vulnerable than others.
The new approach was made possible by the abundance of data related to antibodies against SARS-CoV-2, Wu said.
"In 20 years, scientists have discovered about 5,000 antibodies against the flu virus," he said. "But in just two years, people have identified 8,000 antibodies for COVID. This provides an opportunity that's never been seen before to study how antibodies work and to do this kind of prediction."
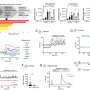
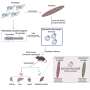
The researchers used antibody data from 88 published studies and 13 patents. The datasets were big enough to allow the researchers to train their model to make predictions based on the antibodies' genetic sequence.
The model was designed to distinguish whether the sequences coded for antibodies targeting regions on the influenza virus or on the SARS-CoV-2 virus. The researchers then checked the accuracy of those predictions.
"The accuracy was close to 85% overall," Wang said.
"I was actually quite surprised that it worked so well," Wu said.
The team is working to improve its model so that it can more precisely determine which parts of the virus the antibodies attack.
"If we can make these predictions based on antibody sequence, we might also be able to go back and designantibodiesthat bind to specificpathogens," Wu said. "This is not something that we can do now, but those are some implications for future study."
More information:Yiquan Wang et al, A large-scale systematic survey reveals recurring molecular features of public antibody responses to SARS-CoV-2,Immunity(2022).DOI: 10.1016/j.immuni.2022.03.019